Products
ChIP™
Given that the discovery of novel, relevant, and safer medicines is often gated by what practicing chemists can and actually do synthesize in the laboratory (i.e. "what can be made" vs. "what should be made"), Eidogen-Sertanty has developed the ChIP system to capture, preserve, and explore medicinally relevant space by delicately balancing synthetic accessibility with medicinal relevancy. Leveraging today's compute power, ChIP will assist in the Prospective Exploration of Synthetically Feasible, Medicinally Relevant Chemical Space (J.Chem. Inf. Model. 2005, 45, 239-248).
ChIP uses a genetic algorithm-driven enumeration engine for automated generation of medicinally relevant, novel molecules with proven synthetic access, facilitating prospective exploration of existing and newly coupled synthetic strategies.
One of the key advantages of ChIP is that it leverages organizationally accessible synthetic methods to drive the enumeration process. By mixing and matching reaction methods, novel compositions of matter are generated with their potential synthetic road-maps.
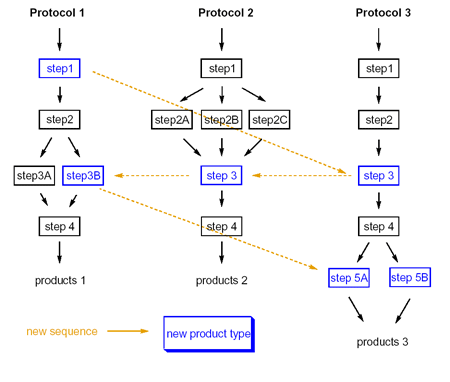
While we offer a starter "basis-set" of reaction chemistries for use with ChIP, the software also readily exploits organizationally accessible, proprietary chemistries captured in the ARK platform. Examples of the ChIP starter reaction basis-set include:
- Subset 1: Nucleophilic Aromatic Substitution Reactions
- Subset 2: Pd-Catalyzed Aromatic Substitutions
- Subset 3: Functional Group Transformations
- Subset 4: Amine Acylation Reactions (Amides / Carbamates)
- Subset 5: Amine Acylation Reactions (Ureas / Thioureas)
- Subset 6: Formation of Diverse Heterocyclic Systems
- Subset 7: Formation of Thioimidazoles
- Subset 8: Standard Deprotection Steps
Download descriptive table of starter reaction chemistry basis-set.
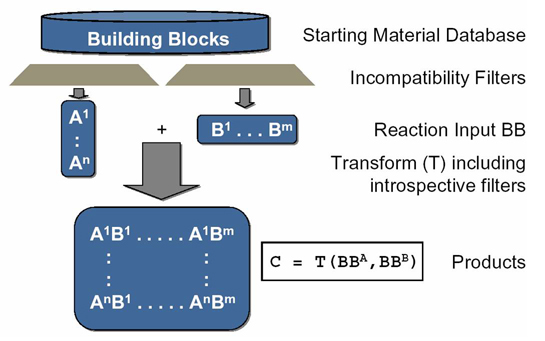
ChIP utilizes a Java™-based, XML-driven command-line enumerator with powerful and useful filtering capabilities to facilitate population of virtual libraries with medicinally relevant, synthetically feasible molecules. Some of ChIP's transform and filtering capabilities are described below:
- Transforms with reactant-specific incompatibility and introspective filters (reaction center environment) defining the scope and limitations of a synthetic reaction step.
- Incompatibility filters (to avoid side reactions) exclude reactive functional groups, e.g. electrophiles, nucleophiles, acids, et c.
- Introspective filters Introspective filters define required reaction center environment (reactivity), e.g. nucleophilic amin, activated aromatic chloride, et c.
- Global "bad-frag" filters avoid reactive functionality or undesired motifs, e.g. acylators, undesired elements/isotopes, et c.
ChIP Applications
- Explore opportunities within your organization's synthetic capabilities and biological know-how.
- Generate biologically relevant molecules that avoid already claimed patent space and meet user-defined filter criteria for potency, selectivity, novelty, eToxicology, or eADME.
- Fill "holes" in structurally diverse compound collections through "mixing-n-matching" of synthetic protocols to generate novel, synthetically accessible molecules.
- Analyze and avoid unproductive library syntheses.
- Predict synthetic feasibility of virtual compounds.
- Improve library synthesis success rates by matching reaction conditions and building blocks.
- Analyze the correlations between building blocks and property distributions of compound libraries.
